Napari Plugin#
CellSAM includes a napari plugin to aid in local, interactive segmentation
of images.
Installation#
The plugin requires napari to be installed.
The simplest way to do so is to install the optional napari dependencies when
installing cellSAM.
For instance, if installing from source in a locally-cloned repo:
pip install .[napari]
Or, to install all necessary dependencies without interacting with the source code:
pip install "cellSAM[napari] @ git+https://github.com/vanvalenlab/cellSAM@master"
Note
The optional dependencies include PyQt6 by default. If you’d like finer-grained control over which Qt bindings to use, consider installing dependencies manually, e.g.:
pip install napari pyside2 magicgui
See the napari documentation for details regarding Qt backends.
Running the plugin#
Open a napari window with the cellSAM plugin:
cellsam napari
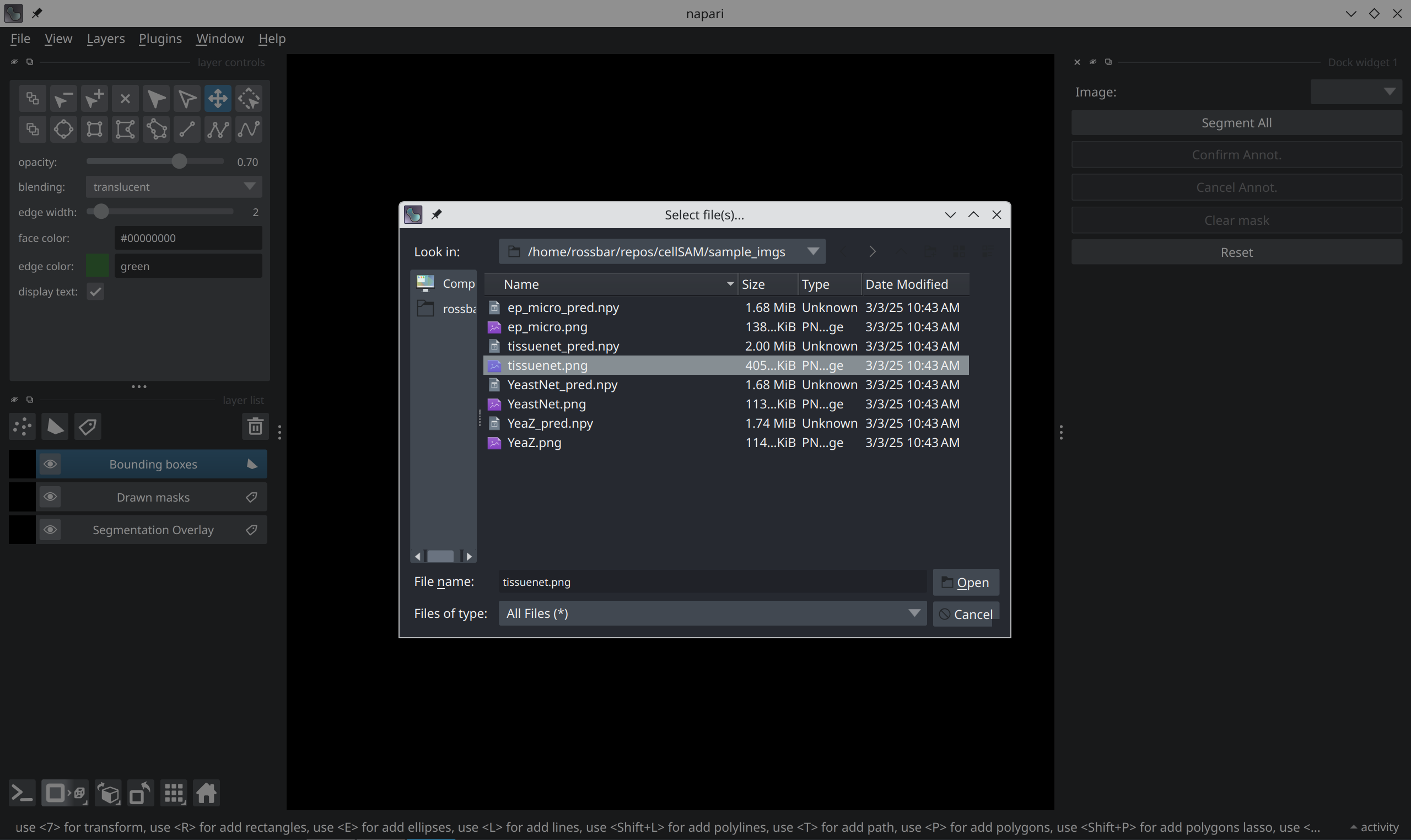
Load an image#
Use the File menu in the upper-right corner and select Open File(s) to
open the loading prompt.
From here, you can select the image you’d like to load — for example,
the tissuenet.png from the sample_imgs/ directory in the source repo:
Note
The plugin currently supports common image formats like .png. Need support for
an additional format? Make a feature request!
Interactive segmentation#
The plugin allows users to specify bounding boxes which prompt the CellSAM model to segment the cell inside the bounding box.
Note
The plugin assumes that there is one cell per bounding box. To perform
segmentation over many cells in an image region, consider the
cellsam_pipeline() library interface.
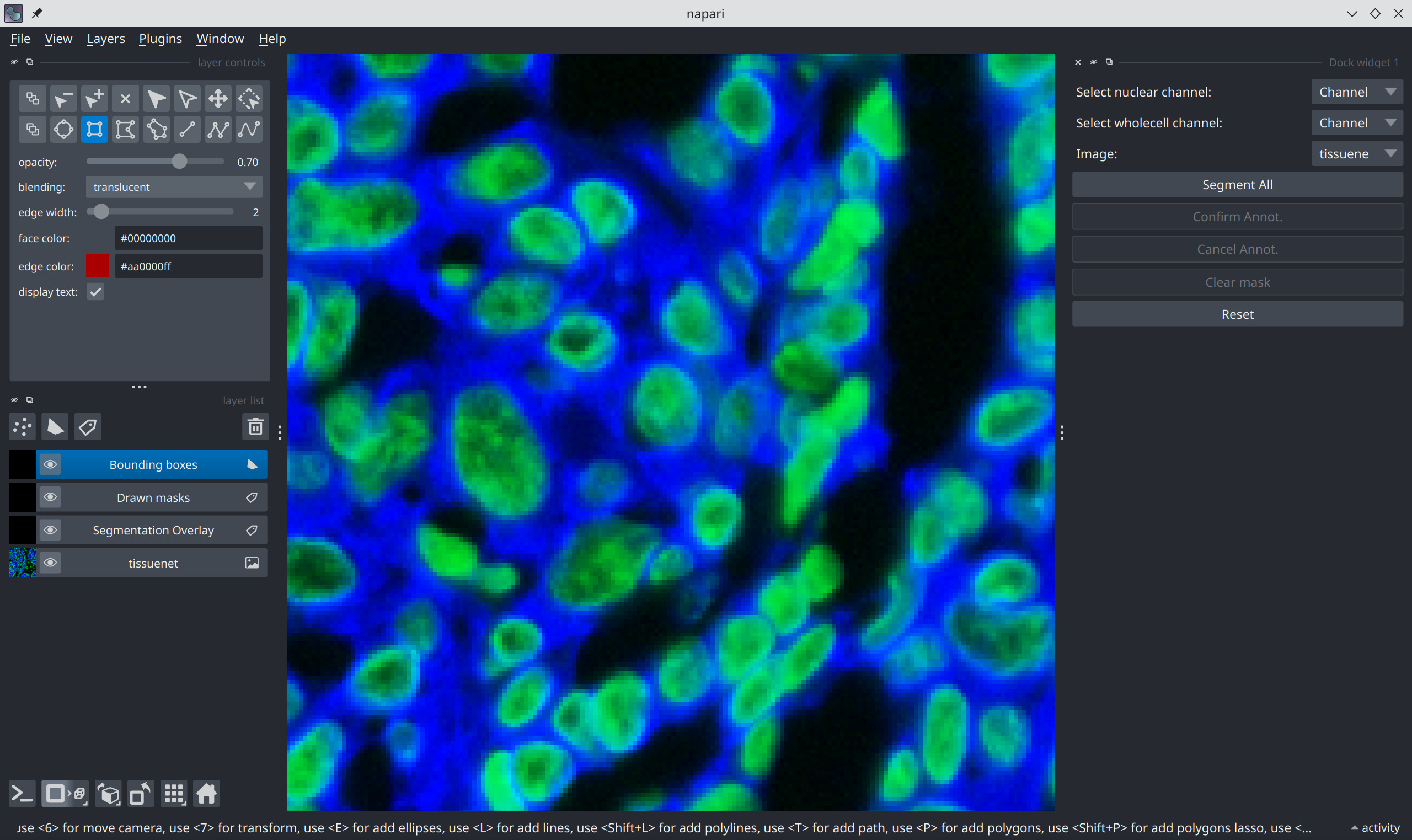
Bounding boxes are drawn by:
Select the “Bounding Boxes” layer on the left panel.
Select the “Add Rectangle” option from the tool panel in the upper left corner:
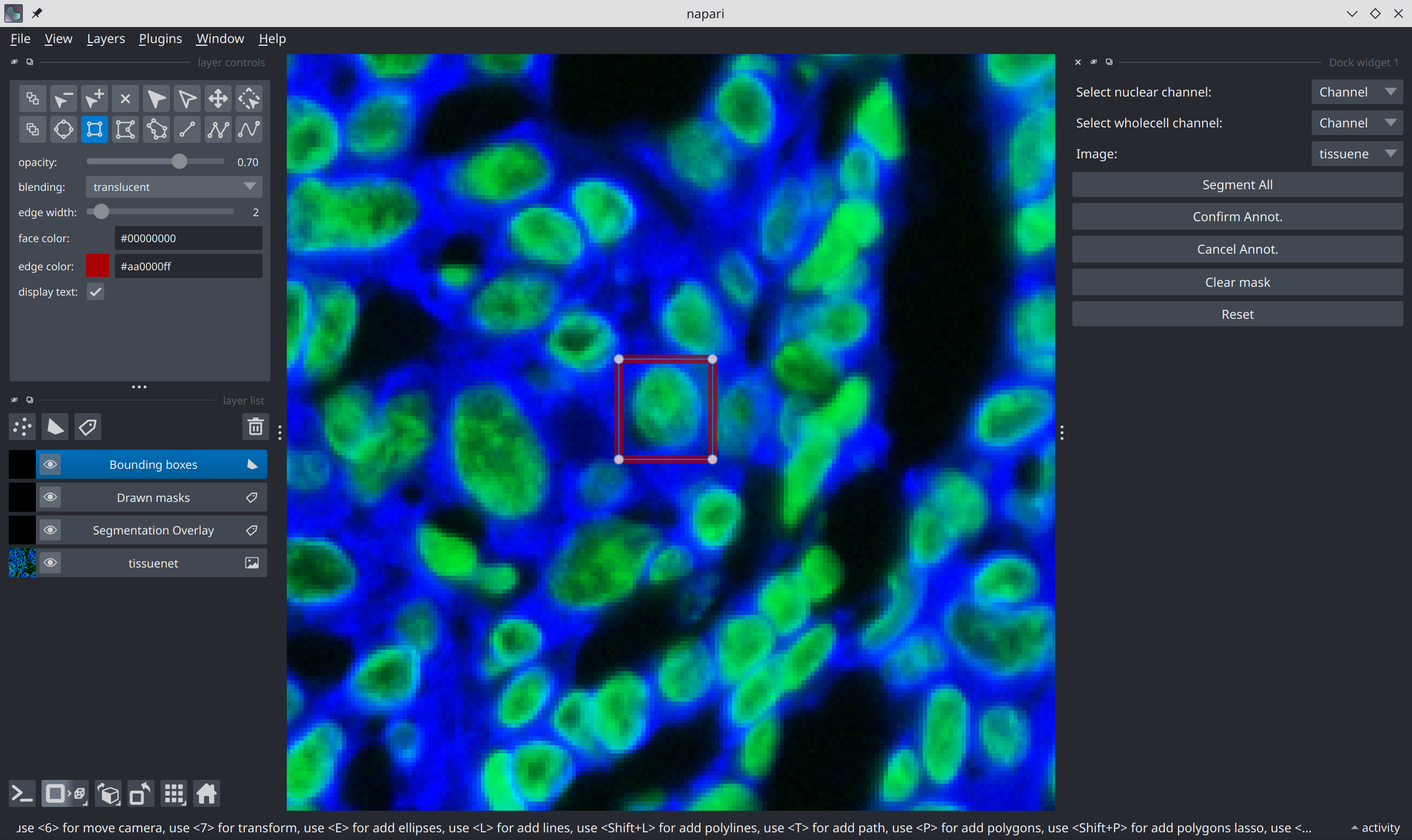
Now, select a region corresponding to a cell you’d like to segment:
Finally, press the “Confirm Annot.” button on the right panel. This will prompt the model to predict the segmentation mask for the highlighted cell.
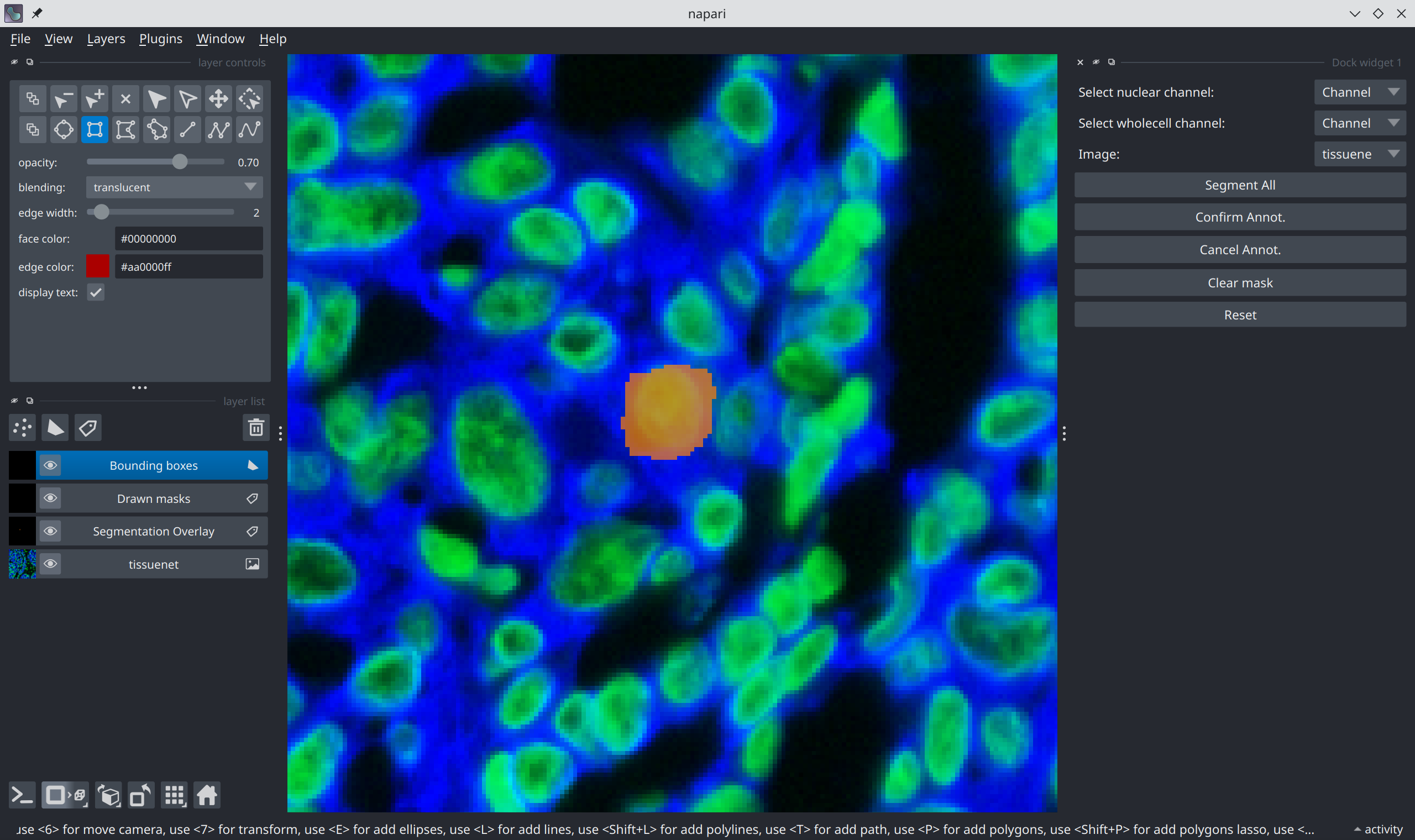
This process can be repeated to segment multiple cells.
The segmented cells will be visible on the main canvas and are captured in the
Segmentation Overlay layer.
Clearing the results#
Individual predictions can be removed using the standard napari labeling tools
in the Segmentation Overlay layer.
The Clear mask button on the right panel can be used to remove all predictions
from the current session (i.e. clear the segmentation mask).
The Reset button removes both the current predictions and the underlying
image and should only be used if you’d like to open a new image in the existing
session.
Segmenting the entire image#
The napari plugin also provides a Segment All button on the right panel which
can be used to automatically segment all cells in the image.
Caution
The napari plugin is primarily designed for interactive segmentation.
The Segment All button is not recommended if:
The image is greater than 2k x 2k pixels.
You do not have a GPU locally available.
The cellsam_pipeline() library interface is
recommended for segmenting entire images.
See the Gallery for examples of how to combine the inference
pipeline with napari visualization.