Note
Go to the end to download the full example code.
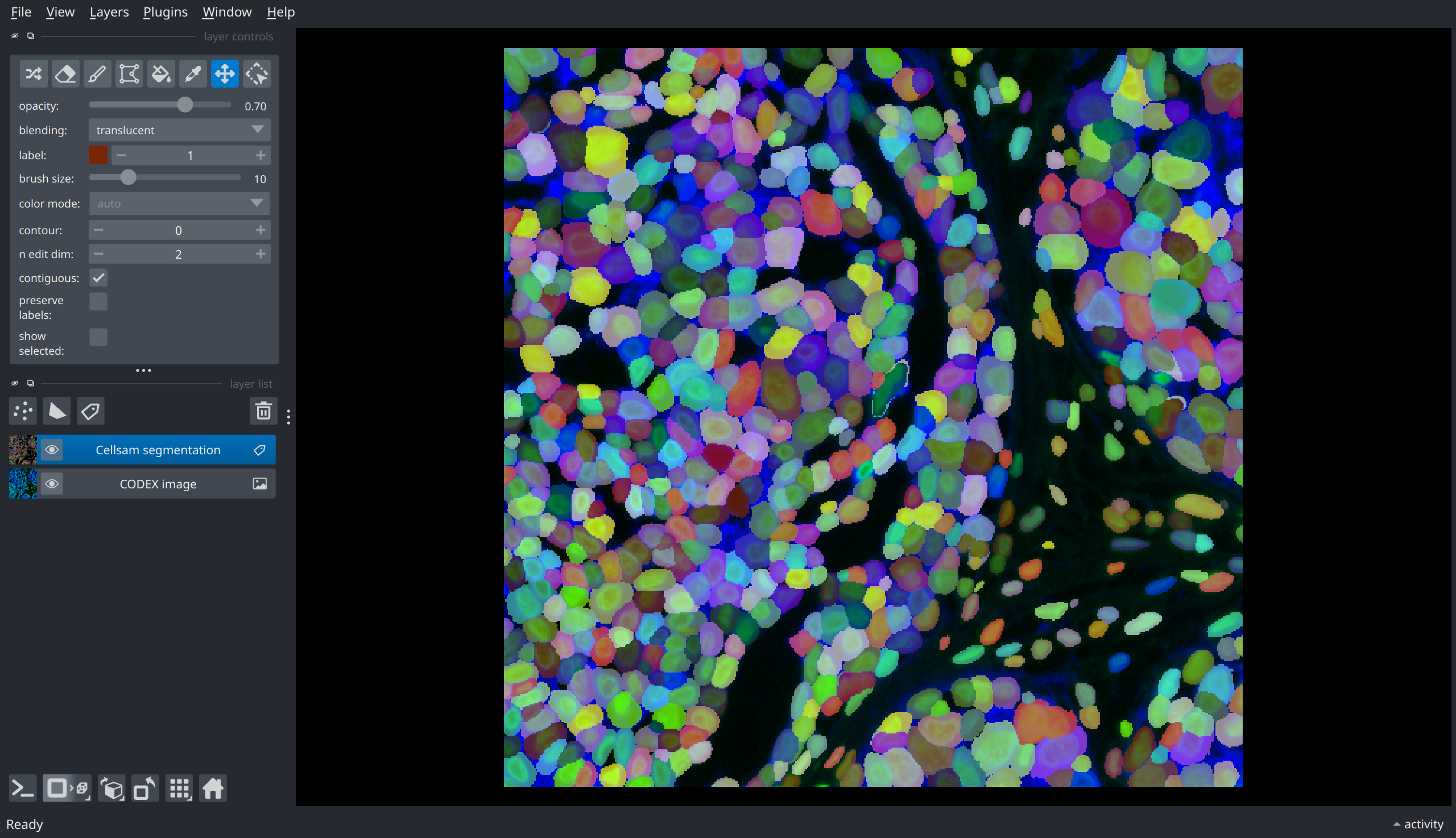
Multiplexed Imaging: CODEX#
[ 0 18481553 27300386]
/home/administrator/envs/cs-13-dev/lib/python3.13/site-packages/cellSAM/sam_inference.py:351: UserWarning: Low IOU threshold, ignoring mask.
warnings.warn("Low IOU threshold, ignoring mask.")
/home/administrator/repos/cellSAM/examples/plot_multiplexed.py:17: FutureWarning: `napari.view_image` is deprecated and will be removed in napari 0.7.0.
Use `viewer = napari.Viewer(); viewer.add_image(...)` instead.
nim = napari.view_image(img, name="CODEX image");
import imageio.v3 as iio
import napari
from cellSAM import cellsam_pipeline
img = iio.imread("../sample_imgs/tissuenet.png")
# Image is 3-channel RGB where Channel 1 (G) represents a nuclear stain
# and Channel 2 (B) a membrane stain. Channel 0 (R) is blank.
print(img.sum(axis=(0, 1)))
mask = cellsam_pipeline(img, use_wsi=False)
nim = napari.view_image(img, name="CODEX image");
nim.add_labels(mask, name="Cellsam segmentation");
if __name__ == "__main__":
napari.run()
Total running time of the script: (0 minutes 7.233 seconds)