Note
Go to the end to download the full example code.
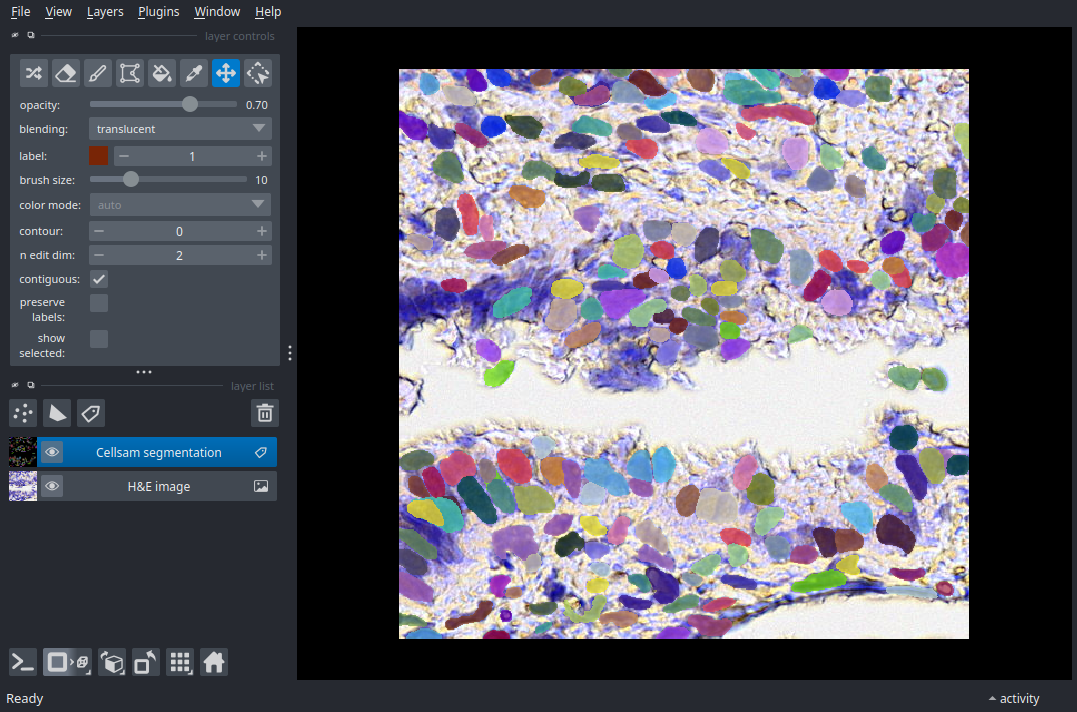
Histology - H&E Staining#
Applying the cellsam_pipeline to segment H&E stained images.
This FOV is extracted from dataset HBM248.QRTB.362 available on the HuBMAP data portal.
(512, 512, 3)
/home/administrator/envs/cs-13-dev/lib/python3.13/site-packages/cellSAM/sam_inference.py:351: UserWarning: Low IOU threshold, ignoring mask.
warnings.warn("Low IOU threshold, ignoring mask.")
/home/administrator/repos/cellSAM/examples/plot_HandE.py:38: FutureWarning: `napari.view_image` is deprecated and will be removed in napari 0.7.0.
Use `viewer = napari.Viewer(); viewer.add_image(...)` instead.
nim = napari.view_image(img, name="H&E image");
import zarr
import skimage
import napari
from cellSAM import cellsam_pipeline
# NOTE: data is stored with zarr_format 3
assert int(zarr.__version__[0]) > 2
# Access H&E image
store = zarr.storage.FsspecStore.from_url(
"s3://cellsam-gallery-sample-data/sample-data.zarr",
storage_options={"anon": True},
read_only=True,
)
z = zarr.open_group(store=store, mode="r")
# Load H&E image into local memory
# Limit to upper-left quadrant to reduce CI computation load
tilesize = 512
img = z["HBM248_QRTB_362"][:tilesize, :tilesize, :]
print(img.shape)
# NOTE: H&E images are often RGB - CellSAM expects RGB images to
# be condensed to a single channel, as with `skimage.color.rgb2gray`,
# for example
mask = cellsam_pipeline(skimage.color.rgb2gray(img), use_wsi=False)
nim = napari.view_image(img, name="H&E image");
nim.add_labels(mask, name="Cellsam segmentation");
if __name__ == "__main__":
napari.run()
Total running time of the script: (0 minutes 5.049 seconds)